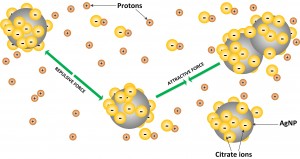
Nanoparticles (NP) undergo interaction and transformation in all media, in vitro or in vivo. Agglomeration, dissolution, reaction, etc. change the number and state of the nanoparticles and thus affect their effect on biological cells. The specific rate and extent of interaction is dependent on the material of the NPs themselves and also on their size, shape, coating, surface zeta potential, and also on the chemical ions present in the media.
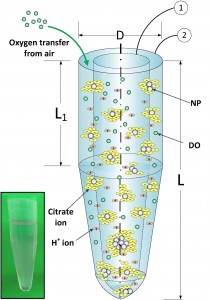
We developed a computational model based on Direct Monte Carlo Simulations (DMCS) which follows the variety of processes taking place in any media. The model called ADSRM (Agglomeration-Dissolution-Sedimentation-Reaction-Model) predicts the state and number of NPs after a certain amount of time in a certain media. The model was implemented for the first time in an Eppendorf tube to compare with in vitro measurements.
ADSRM can be implemented in a variety of media and a variety of geometries and can be used to predict NP dosimetry to cells and tissues to understand cellular uptake, and NP toxicology.
For further details of the ADSRM model, the published article can be accessed here:
Mukherjee et al., Journal of Nanoparticle Research, 2014, 16, 2616.